Compound ccmrd_632
Compound Info
| Database id | Trival name | Linear code | Compound class | Taxonomy domain | Species |
|---|---|---|---|---|---|
| ccmrd_632 | N-acetylmuramic acid | MurpNAc | peptidoglycan | bacteria | Bacillus subtilis |
Residues
| # | Residue | Chemical Structure | Chemical Shifts (ppm) |
|---|---|---|---|
| 1 | ?-?-MurpNAc | 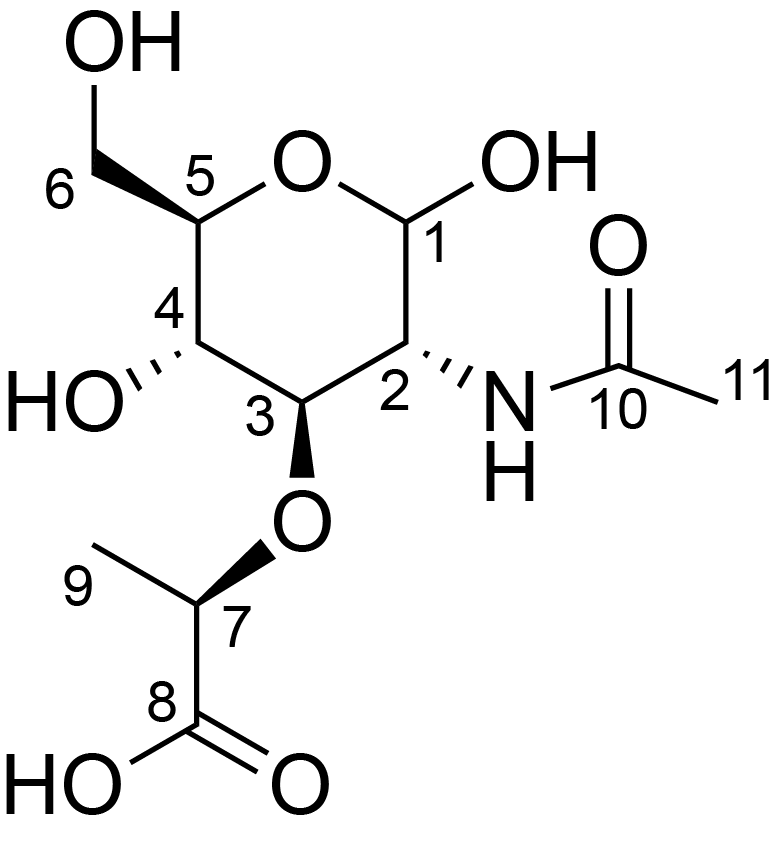 |
C 1 : 103.6 ; C 2 : 56.4 ; C 3 : 80.6 ; C 4 : 73.7 ; C 5 : 76.0 ; C 6 : 78.6 ; C 7 : 61.1 ; C 8 : ; C 9 : 18.4 ; C 10 : ; C 11 : 23.1 ; H 1 : 4.4 ; H 2 : 3.7 ; H 3 : 3.5 ; H 4 : 3.3 ; H 5 : 3.8 ; H 6 : 3.7 ; H 7 : 4.4 ; H 8 : ; H 9 : 1.3 ; H 10 : ; H 11 : 1.9 ; H 12 : 7.8 ; N 1 : 122.6 ; |
Experiment
| Spectrometer (MHz) | Temperature (K) | pH | NMR Reference Compound | Sample treatment |
|---|---|---|---|---|
| 950 | 303 | TMS | Extracting intact well-hydrated peptidoglycan sacculi |
Reference
Catherine Bougault,
Isabel Ayala,
Waldemar Vollmer,
Jean-Pierre Simorre,
Paul Schanda,
(2019). Studying intact bacterial peptidoglycan by proton-detected NMR spectroscopy at 100 kHz MAS frequency,
Journal of Structural Biology,
206
(1)
,66-72
.
doi:10.1016/j.jsb.2018.07.009